Parametric Classification Models#
Machine Learning Methods#
Module 4: Parametric Classification Models#
Instructor: Farhad Pourkamali#
Overview#
Logistic regression: Problem formulation, assumption, loss function, gradient
Logistic regression in Scikit-learn (sklearn)
Video for parts 1 and 2: https://youtu.be/N_4nVvcYQHo
Evaluation metrics: https://youtu.be/vzv4Q7Fq98s
Multiclass classification: https://youtu.be/UoN7O4cJat0
1. Foundations of Logistic Regression#
Logistic regression#
Logistic regression involves a probabilistic model of the form \(p(y|\mathbf{x};\boldsymbol{\theta})\), where \(\mathbf{x}\in\mathbb{R}^D\) is a fixed-dimensional input vector
\(C=2\): binary logistic regression \(\rightarrow y\in\{0,1\}\)
\(C>2\): multinomial/multiclass logistic regression \(\rightarrow y\in\{1,2,\ldots,C\}\)
Recall the probability mass function (pmf) of the Bernoulli distribution
Binary logistic regression
The function \(\color{red}{f(\mathbf{x}; \boldsymbol{\theta})}\) is a linear combination of the input features, i.e., \(f(\mathbf{x}; \boldsymbol{\theta}) = \mathbf{x}^\top \boldsymbol{\theta}\), which is transformed by the sigmoid function \(\color{red}{\sigma(a) = \frac{1}{1 + e^{-a}}}\) to map the linear output into a probability space
import numpy as np
import matplotlib.pyplot as plt
plt.rcParams.update({'font.size': 12, "figure.figsize": (5,3)})
a = np.linspace(-10, 10, 100)
sig = 1 / (1 + np.exp(-a))
plt.plot(a, sig, "b-", linewidth=2, label=r"$\sigma(a) = \frac{1}{1 + e^{-a}}$")
plt.xlabel("a")
plt.legend(loc="upper left")
plt.axis([-10, 10, -0.1, 1.1])
plt.grid(color='0.95')
plt.show()
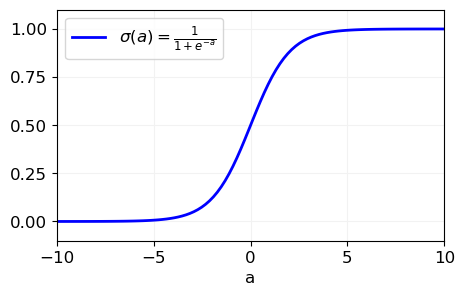
Properties of sigmoid function#
The derivative of \(\sigma(a)\) has a nice form
a = np.linspace(-10, 10, 100)
sig = 1 / (1 + np.exp(-a))
plt.plot(a, sig, "b-", linewidth=2, label=r"$\sigma(a) $")
plt.plot(a, sig*(1-sig), "r-", linewidth=2, label=r"$\sigma(a)'$")
plt.xlabel("a")
plt.legend(loc="upper left")
plt.axis([-10, 10, -0.1, 1.1])
plt.grid(color='0.95')
plt.show()
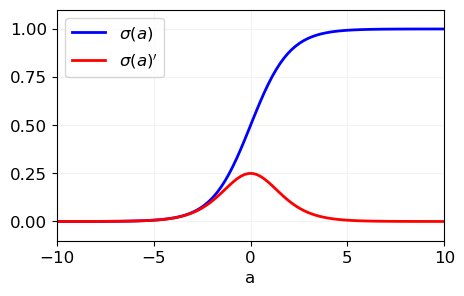
Binary classification#
Plugging the definition of the sigmoid function
The quantity \(a\) is known as the log-odds or logit
The inverse of the sigmoid function is called the logit function
Hence, the sigmoid (or logistic) function is an invertible function that allows us to:
Convert a continuous number \(a\) into the probability space \([0, 1]\) using \(\sigma(a)\)
Convert a probability back into the corresponding log-odds using the logit function \(\text{logit}(p)\)
import numpy as np
from scipy.special import expit, logit
import matplotlib.pyplot as plt
fig, ax = plt.subplots(1,2, figsize=(7,3))
# expit
x = np.linspace(-5, 5, 100)
ax[0].plot(x, expit(x), 'r')
ax[0].set_xlabel('a')
ax[0].set_ylabel('p')
ax[0].grid()
ax[0].set_title('logistic')
# logit
x = np.linspace(0, 1, 100)
ax[1].plot(x, logit(x), 'r')
ax[1].set_xlabel('p')
ax[1].set_ylabel('a')
ax[1].grid()
ax[1].set_title('logit')
plt.tight_layout()
plt.show()
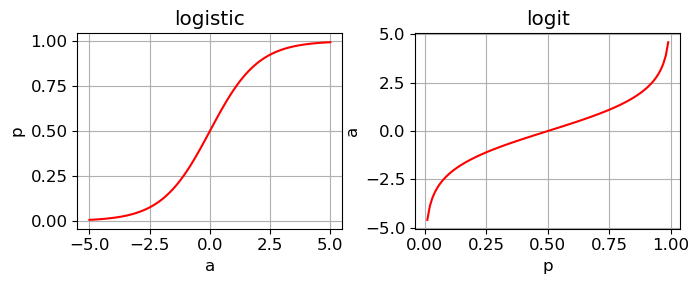
Linear model for binary logistic regression#
Use a linear function of the form \(f(\mathbf{x};\boldsymbol{\theta})=\boldsymbol{\theta}^T\mathbf{x}\), yielding the following pmf
Thus, we get
Optimal Decision Rule for Predicting \(y=1\)#
Starting point - Probability comparison:
The optimal decision rule is based on comparing the probabilities of the two classes. To predict \(y=1\), we check if:\[p(y=1|\mathbf{x}) > p(y=0|\mathbf{x})\]Log-odds transformation:
Using the definition of log-odds (logit), the inequality can be rewritten as:\[\log \frac{p(y=1|\mathbf{x})}{p(y=0|\mathbf{x})} > 0\]This shows that predicting \(y=1\) is equivalent to determining whether the log-odds are positive
Linear form of log-odds:
For logistic regression, the log-odds are given by:\[\log \frac{p(y=1|\mathbf{x})}{p(y=0|\mathbf{x})} = \boldsymbol{\theta}^\top \mathbf{x}\]Thus, the decision rule reduces to:
\[\boldsymbol{\theta}^\top \mathbf{x} > 0.\]Interpretation of the rule:
If \(\boldsymbol{\theta}^\top \mathbf{x} > 0\), we predict \(y=1\)
If \(\boldsymbol{\theta}^\top \mathbf{x} \leq 0\), we predict \(y=0\)
This shows that the decision boundary is defined by the hyperplane \(\boldsymbol{\theta}^\top \mathbf{x} = 0\), which separates the feature space into two regions
Geometric intuition:
The vector \(\boldsymbol{\theta}\) determines the orientation of the decision boundary in the feature space
The magnitude of \(\boldsymbol{\theta}^\top \mathbf{x}\) indicates how confidently a sample is classified, with larger values implying higher certainty
Loss function#
Logistic regression model estimates probabilities and makes predictions. But how is it trained?
Let us define \(\mu_n=\sigma(a_n)\) and \(a_n=\boldsymbol{\theta}^T\mathbf{x}_n\)
The loss function \(l(y_n, \mu_n)\) quantifies the error between the true label \(y_n\) and the predicted probability \(\mu_n\)
When the true label \(y_n=1\), the model is penalized based on the negative logarithm of the predicted probability \(\mu_n\)
If \(\mu_n\) is close to 1, the penalty is small; if it’s close to 0, the penalty is large
When \(y_n=0\), the model is penalized based on the negative logarithm of \(1-\mu_n\), which represents the predicted probability of \(y_n=0\)
Essentially, the loss is the negative log of the probability assigned to the correct class
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from scipy.special import expit
def cross_entropy_loss(y, mu):
if y == 1:
return -np.log(mu)
else:
return -np.log(1 - mu)
z = np.arange(-10, 10, 0.1)
mu_z = expit(z)
cost_1 = cross_entropy_loss(1, mu_z) # when y = 1
cost_0 = cross_entropy_loss(0, mu_z) # when y = 0
fig, ax = plt.subplots(figsize=(4,3))
plt.plot(mu_z, cost_1, 'C4-', label='y=1')
plt.plot(mu_z, cost_0, 'C8-', label='y=0')
plt.xlabel('prob(y=1)')
plt.ylabel('loss')
plt.tight_layout()
plt.legend()
plt.grid()
plt.show()
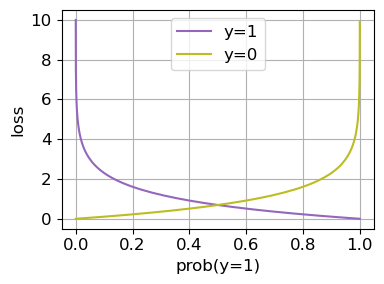
Loss function#
The loss function over the whole training set is the average loss over all training samples
Can we compute the gradient? Recall the data matrix \(\mathbf{X}\in\mathbb{R}^{N\times D}\) and target vector \(\mathbf{y}\in\mathbb{R}^N\)
Thus, partial derivatives can be written as
2. Logistic regression in Scikit-learn (sklearn)#
from sklearn.datasets import load_iris
iris = load_iris(as_frame=True)
list(iris)
['data',
'target',
'frame',
'target_names',
'DESCR',
'feature_names',
'filename',
'data_module']
iris['data']
| sepal length (cm) | sepal width (cm) | petal length (cm) | petal width (cm) | |
|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 |
| ... | ... | ... | ... | ... |
| 145 | 6.7 | 3.0 | 5.2 | 2.3 |
| 146 | 6.3 | 2.5 | 5.0 | 1.9 |
| 147 | 6.5 | 3.0 | 5.2 | 2.0 |
| 148 | 6.2 | 3.4 | 5.4 | 2.3 |
| 149 | 5.9 | 3.0 | 5.1 | 1.8 |
150 rows × 4 columns
iris['target'].value_counts(normalize=True)
target
0 0.333333
1 0.333333
2 0.333333
Name: proportion, dtype: float64
iris['target_names']
array(['setosa', 'versicolor', 'virginica'], dtype='<U10')
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import train_test_split
X = iris.data[["petal width (cm)"]].values
y = iris.target_names[iris.target] == 'virginica'
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=42)
log_reg = LogisticRegression(random_state=42)
log_reg.fit(X_train, y_train)
LogisticRegression(random_state=42)In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
LogisticRegression(random_state=42)
y
array([False, False, False, False, False, False, False, False, False,
False, False, False, False, False, False, False, False, False,
False, False, False, False, False, False, False, False, False,
False, False, False, False, False, False, False, False, False,
False, False, False, False, False, False, False, False, False,
False, False, False, False, False, False, False, False, False,
False, False, False, False, False, False, False, False, False,
False, False, False, False, False, False, False, False, False,
False, False, False, False, False, False, False, False, False,
False, False, False, False, False, False, False, False, False,
False, False, False, False, False, False, False, False, False,
False, True, True, True, True, True, True, True, True,
True, True, True, True, True, True, True, True, True,
True, True, True, True, True, True, True, True, True,
True, True, True, True, True, True, True, True, True,
True, True, True, True, True, True, True, True, True,
True, True, True, True, True, True])
X_new = np.linspace(0, 3, 1000).reshape(-1, 1) # reshape to get a column vector
y_proba = log_reg.predict_proba(X_new)
print(y_proba)
[[0.99820801 0.00179199]
[0.99818732 0.00181268]
[0.99816638 0.00183362]
...
[0.00578965 0.99421035]
[0.00572381 0.99427619]
[0.00565872 0.99434128]]
X_new[y_proba[:, 1] >= 0.5]
array([[1.65165165],
[1.65465465],
[1.65765766],
[1.66066066],
[1.66366366],
[1.66666667],
[1.66966967],
[1.67267267],
[1.67567568],
[1.67867868],
[1.68168168],
[1.68468468],
[1.68768769],
[1.69069069],
[1.69369369],
[1.6966967 ],
[1.6996997 ],
[1.7027027 ],
[1.70570571],
[1.70870871],
[1.71171171],
[1.71471471],
[1.71771772],
[1.72072072],
[1.72372372],
[1.72672673],
[1.72972973],
[1.73273273],
[1.73573574],
[1.73873874],
[1.74174174],
[1.74474474],
[1.74774775],
[1.75075075],
[1.75375375],
[1.75675676],
[1.75975976],
[1.76276276],
[1.76576577],
[1.76876877],
[1.77177177],
[1.77477477],
[1.77777778],
[1.78078078],
[1.78378378],
[1.78678679],
[1.78978979],
[1.79279279],
[1.7957958 ],
[1.7987988 ],
[1.8018018 ],
[1.8048048 ],
[1.80780781],
[1.81081081],
[1.81381381],
[1.81681682],
[1.81981982],
[1.82282282],
[1.82582583],
[1.82882883],
[1.83183183],
[1.83483483],
[1.83783784],
[1.84084084],
[1.84384384],
[1.84684685],
[1.84984985],
[1.85285285],
[1.85585586],
[1.85885886],
[1.86186186],
[1.86486486],
[1.86786787],
[1.87087087],
[1.87387387],
[1.87687688],
[1.87987988],
[1.88288288],
[1.88588589],
[1.88888889],
[1.89189189],
[1.89489489],
[1.8978979 ],
[1.9009009 ],
[1.9039039 ],
[1.90690691],
[1.90990991],
[1.91291291],
[1.91591592],
[1.91891892],
[1.92192192],
[1.92492492],
[1.92792793],
[1.93093093],
[1.93393393],
[1.93693694],
[1.93993994],
[1.94294294],
[1.94594595],
[1.94894895],
[1.95195195],
[1.95495495],
[1.95795796],
[1.96096096],
[1.96396396],
[1.96696697],
[1.96996997],
[1.97297297],
[1.97597598],
[1.97897898],
[1.98198198],
[1.98498498],
[1.98798799],
[1.99099099],
[1.99399399],
[1.996997 ],
[2. ],
[2.003003 ],
[2.00600601],
[2.00900901],
[2.01201201],
[2.01501502],
[2.01801802],
[2.02102102],
[2.02402402],
[2.02702703],
[2.03003003],
[2.03303303],
[2.03603604],
[2.03903904],
[2.04204204],
[2.04504505],
[2.04804805],
[2.05105105],
[2.05405405],
[2.05705706],
[2.06006006],
[2.06306306],
[2.06606607],
[2.06906907],
[2.07207207],
[2.07507508],
[2.07807808],
[2.08108108],
[2.08408408],
[2.08708709],
[2.09009009],
[2.09309309],
[2.0960961 ],
[2.0990991 ],
[2.1021021 ],
[2.10510511],
[2.10810811],
[2.11111111],
[2.11411411],
[2.11711712],
[2.12012012],
[2.12312312],
[2.12612613],
[2.12912913],
[2.13213213],
[2.13513514],
[2.13813814],
[2.14114114],
[2.14414414],
[2.14714715],
[2.15015015],
[2.15315315],
[2.15615616],
[2.15915916],
[2.16216216],
[2.16516517],
[2.16816817],
[2.17117117],
[2.17417417],
[2.17717718],
[2.18018018],
[2.18318318],
[2.18618619],
[2.18918919],
[2.19219219],
[2.1951952 ],
[2.1981982 ],
[2.2012012 ],
[2.2042042 ],
[2.20720721],
[2.21021021],
[2.21321321],
[2.21621622],
[2.21921922],
[2.22222222],
[2.22522523],
[2.22822823],
[2.23123123],
[2.23423423],
[2.23723724],
[2.24024024],
[2.24324324],
[2.24624625],
[2.24924925],
[2.25225225],
[2.25525526],
[2.25825826],
[2.26126126],
[2.26426426],
[2.26726727],
[2.27027027],
[2.27327327],
[2.27627628],
[2.27927928],
[2.28228228],
[2.28528529],
[2.28828829],
[2.29129129],
[2.29429429],
[2.2972973 ],
[2.3003003 ],
[2.3033033 ],
[2.30630631],
[2.30930931],
[2.31231231],
[2.31531532],
[2.31831832],
[2.32132132],
[2.32432432],
[2.32732733],
[2.33033033],
[2.33333333],
[2.33633634],
[2.33933934],
[2.34234234],
[2.34534535],
[2.34834835],
[2.35135135],
[2.35435435],
[2.35735736],
[2.36036036],
[2.36336336],
[2.36636637],
[2.36936937],
[2.37237237],
[2.37537538],
[2.37837838],
[2.38138138],
[2.38438438],
[2.38738739],
[2.39039039],
[2.39339339],
[2.3963964 ],
[2.3993994 ],
[2.4024024 ],
[2.40540541],
[2.40840841],
[2.41141141],
[2.41441441],
[2.41741742],
[2.42042042],
[2.42342342],
[2.42642643],
[2.42942943],
[2.43243243],
[2.43543544],
[2.43843844],
[2.44144144],
[2.44444444],
[2.44744745],
[2.45045045],
[2.45345345],
[2.45645646],
[2.45945946],
[2.46246246],
[2.46546547],
[2.46846847],
[2.47147147],
[2.47447447],
[2.47747748],
[2.48048048],
[2.48348348],
[2.48648649],
[2.48948949],
[2.49249249],
[2.4954955 ],
[2.4984985 ],
[2.5015015 ],
[2.5045045 ],
[2.50750751],
[2.51051051],
[2.51351351],
[2.51651652],
[2.51951952],
[2.52252252],
[2.52552553],
[2.52852853],
[2.53153153],
[2.53453453],
[2.53753754],
[2.54054054],
[2.54354354],
[2.54654655],
[2.54954955],
[2.55255255],
[2.55555556],
[2.55855856],
[2.56156156],
[2.56456456],
[2.56756757],
[2.57057057],
[2.57357357],
[2.57657658],
[2.57957958],
[2.58258258],
[2.58558559],
[2.58858859],
[2.59159159],
[2.59459459],
[2.5975976 ],
[2.6006006 ],
[2.6036036 ],
[2.60660661],
[2.60960961],
[2.61261261],
[2.61561562],
[2.61861862],
[2.62162162],
[2.62462462],
[2.62762763],
[2.63063063],
[2.63363363],
[2.63663664],
[2.63963964],
[2.64264264],
[2.64564565],
[2.64864865],
[2.65165165],
[2.65465465],
[2.65765766],
[2.66066066],
[2.66366366],
[2.66666667],
[2.66966967],
[2.67267267],
[2.67567568],
[2.67867868],
[2.68168168],
[2.68468468],
[2.68768769],
[2.69069069],
[2.69369369],
[2.6966967 ],
[2.6996997 ],
[2.7027027 ],
[2.70570571],
[2.70870871],
[2.71171171],
[2.71471471],
[2.71771772],
[2.72072072],
[2.72372372],
[2.72672673],
[2.72972973],
[2.73273273],
[2.73573574],
[2.73873874],
[2.74174174],
[2.74474474],
[2.74774775],
[2.75075075],
[2.75375375],
[2.75675676],
[2.75975976],
[2.76276276],
[2.76576577],
[2.76876877],
[2.77177177],
[2.77477477],
[2.77777778],
[2.78078078],
[2.78378378],
[2.78678679],
[2.78978979],
[2.79279279],
[2.7957958 ],
[2.7987988 ],
[2.8018018 ],
[2.8048048 ],
[2.80780781],
[2.81081081],
[2.81381381],
[2.81681682],
[2.81981982],
[2.82282282],
[2.82582583],
[2.82882883],
[2.83183183],
[2.83483483],
[2.83783784],
[2.84084084],
[2.84384384],
[2.84684685],
[2.84984985],
[2.85285285],
[2.85585586],
[2.85885886],
[2.86186186],
[2.86486486],
[2.86786787],
[2.87087087],
[2.87387387],
[2.87687688],
[2.87987988],
[2.88288288],
[2.88588589],
[2.88888889],
[2.89189189],
[2.89489489],
[2.8978979 ],
[2.9009009 ],
[2.9039039 ],
[2.90690691],
[2.90990991],
[2.91291291],
[2.91591592],
[2.91891892],
[2.92192192],
[2.92492492],
[2.92792793],
[2.93093093],
[2.93393393],
[2.93693694],
[2.93993994],
[2.94294294],
[2.94594595],
[2.94894895],
[2.95195195],
[2.95495495],
[2.95795796],
[2.96096096],
[2.96396396],
[2.96696697],
[2.96996997],
[2.97297297],
[2.97597598],
[2.97897898],
[2.98198198],
[2.98498498],
[2.98798799],
[2.99099099],
[2.99399399],
[2.996997 ],
[3. ]])
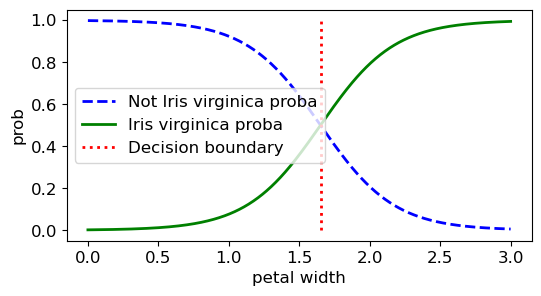
decision_boundary = X_new[y_proba[:, 1] >= 0.5][0, 0]
plt.figure(figsize=(6, 3))
plt.plot(X_new, y_proba[:, 0], "b--", linewidth=2,
label="Not Iris virginica proba")
plt.plot(X_new, y_proba[:, 1], "g-", linewidth=2, label="Iris virginica proba")
plt.plot([decision_boundary, decision_boundary], [0, 1], "r:", linewidth=2,
label="Decision boundary")
plt.xlabel('petal width')
plt.ylabel('prob')
plt.legend()
plt.show()
log_reg.predict_proba(X_test[:5])
array([[0.84890373, 0.15109627],
[0.99436711, 0.00563289],
[0.0767371 , 0.9232629 ],
[0.6403484 , 0.3596516 ],
[0.72310724, 0.27689276]])
log_reg.predict(X_test[:5])
array([False, False, True, False, False])
3. Evaluation metrics#
MNIST is one of the most popular benchmark data sets in machine learning, which can be accessed via scikit-learn
Thus, it involves minimal to no preprocessing
70,000 images, each labeled with the digit it represents
Each image is 28 x 28 pixels, i.e., a 2D array, but stored as a 1D array with 784 features
Note that \(28^2=784\)
Each feature shows the intensity of one pixel
The grayscale intensity values between 0 and 255, corresponding to shades of gray
0 being the lightest (white) and 255 being the darkest (black)
Classifier: https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.SGDClassifier.html
import numpy as np
from sklearn.datasets import fetch_openml
mnist = fetch_openml('mnist_784', version=1, as_frame=False)
mnist.keys()
dict_keys(['data', 'target', 'frame', 'categories', 'feature_names', 'target_names', 'DESCR', 'details', 'url'])
mnist['data'].shape # 70,000 images in R^784
(70000, 784)
mnist['target'] # 70,000 labels
array(['5', '0', '4', ..., '4', '5', '6'], dtype=object)
# Identify input features and labels
X, y = mnist.data, mnist.target
# Let us look at the label of this image and its type
print(y[0],'\n', type(y[0]))
5
<class 'str'>
# train/test split
X_train, X_test, y_train, y_test = X[:60000], X[60000:], y[:60000], y[60000:]
X_train.shape, X_test.shape
((60000, 784), (10000, 784))
# create a binary classification problem (detecting digit 5)
y_train_5 = (y_train == '5')
y_test_5 = (y_test == '5')
y_train_5[:10]
array([ True, False, False, False, False, False, False, False, False,
False])
# We select a classifier and measure its "accuracy" using cross validation
from sklearn.linear_model import SGDClassifier
from sklearn.model_selection import cross_val_score
sgd_clf = SGDClassifier(loss='log_loss', max_iter=1000, tol=1e-3, random_state=42)
# accuracy: the fraction of correct predictions over the total number of samples
cross_val_score(sgd_clf, X_train, y_train_5, cv=3, scoring="accuracy")
array([0.9687 , 0.9616 , 0.96165])
# Let us investigate this "amazing" result more by defining a
# classifier that always returns False
from sklearn.base import BaseEstimator
class Never5Classifier(BaseEstimator):
def fit(self, X, y=None):
pass # no training
def predict(self, X):
return np.zeros((len(X), 1), dtype=bool) # return False for every input image
cross_val_score(Never5Classifier(), X_train, y_train_5, cv=3, scoring="accuracy")
array([0.91125, 0.90855, 0.90915])
# In this cell, we investigate one of the most popular ways of evaluating classifiers
sgd_clf = SGDClassifier(loss='log_loss', max_iter=1000, tol=1e-3, random_state=42)
sgd_clf.fit(X_train, y_train_5)
y_pred_5 = sgd_clf.predict(X_test)
import matplotlib.pyplot as plt
from sklearn.metrics import ConfusionMatrixDisplay
plt.rcParams.update({'font.size': 14, "figure.figsize": (5,3)})
ConfusionMatrixDisplay.from_predictions(y_test_5, y_pred_5, display_labels=np.array([-1,1]))
plt.show()
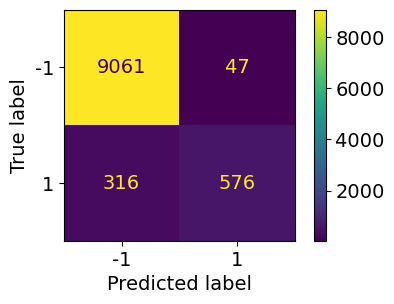
Confusion matrix#
Each row in a confusion matrix represents an actual class, while each column represents a predicted class
The first row of this matrix considers non-5 images (negative class)
The second row considers the images of 5s (positive class)

from sklearn.metrics import precision_score, recall_score
print("Precision: %0.2f" %precision_score(y_test_5, y_pred_5))
print("Recall: %0.2f" %recall_score(y_test_5, y_pred_5))
Precision: 0.92
Recall: 0.65
Precision and recall#
We can combine precision and recall into a single metric called the \(F_1\) score using the harmonic mean
A classifier gets a high \(F_1\) score if both recall and precision are high
Also, we can plot precision/recall values as a function of the threshold used for classification
Most classifiers, such as logistic regression or deep networks, output a score \(S(\mathbf{x})=P(y=1 | \mathbf{x})\) that represents the probability (or confidence) of a sample being in the positive class:
To make a classification decision, we set a threshold \(\tau\) and classify a sample as positive if:
and negative otherwise
High Threshold \(\tau\) (e.g., 0.9): Only very confident positive predictions are classified as positive → High precision, low recall
Low Threshold \(\tau\) (e.g., 0.1): Many uncertain cases are classified as positive → High recall, but more false positives (lower precision)
from sklearn.metrics import PrecisionRecallDisplay
plt.rcParams.update({'font.size': 12, "figure.figsize": (7,6)})
# Use the fitted classifier sgd_clf
PrecisionRecallDisplay.from_estimator(sgd_clf, X_test, y_test_5)
# Previous results
plt.scatter(recall_score(y_test_5, y_pred_5), precision_score(y_test_5, y_pred_5), c = 'r', s=50)
plt.grid()
plt.show()
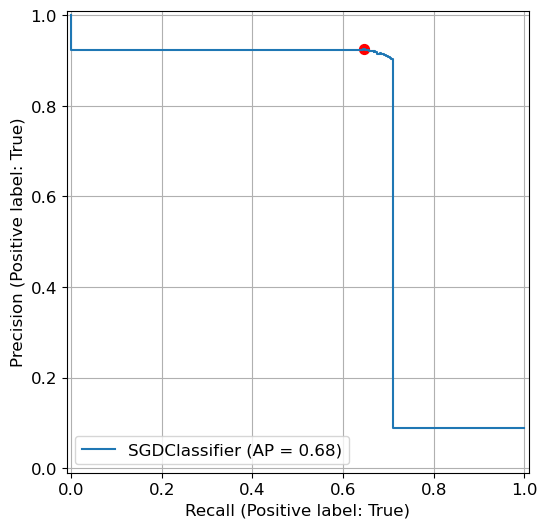
What Does AP Measure?
AP measures the area under the PR curve by summing the contribution of precision values at different recall levels
Instead of computing the exact integral under the curve (which is continuous), AP uses a discrete approximation by dividing the area into small trapezoidal sections
Where:
\(P_i\) is the precision at recall \(R_i\)
\(R_i - R_{i-1}\) is the change in recall (width of the step)
The summation approximates the integral of precision over recall
4. Multiclass classification#
Multinomial Logistic Regression#
Generalizing binary classification to multiple classes:
To represent a distribution over \(C\) possible classes \(y \in \{1, \ldots, C\}\), we use the categorical distribution, which generalizes the Bernoulli distribution for \(C > 2\). The categorical distribution is given by: $\(\text{Cat}(y|\boldsymbol{\theta}) = \prod_{c=1}^C \theta_c^{I(y=c)}\)$ where:\(\boldsymbol{\theta} = [\theta_1, \ldots, \theta_C]\) is the probability vector such that \(0 \leq \theta_c \leq 1\) and \(\sum_{c=1}^C \theta_c = 1\)
\(I(y=c)\) is an indicator function that equals 1 if \(y = c\), and 0 otherwise
Modeling class probabilities:
A multinomial logistic regression model represents the probability of class \(y\) as:\[p(y|\mathbf{x}; \boldsymbol{\theta}) = \text{Cat}\big(y|\color{red}{\text{softmax}\big(f(\mathbf{x}; \boldsymbol{\theta})\big)}\big)\]Softmax transformation:
Let \(\mathbf{a} = \mathbf{W}\mathbf{x} \in \mathbb{R}^C\) represent the logits (the unnormalized scores for each class)
The softmax function transforms the logits into a valid probability distribution over \(C\) classes:
\[\boldsymbol{\theta} = \text{softmax}(\mathbf{a}) = \Big[\frac{e^{a_1}}{\sum_{c'=1}^C e^{a_{c'}}}, \ldots, \frac{e^{a_C}}{\sum_{c'=1}^C e^{a_{c'}}}\Big]\]
Cross-entropy loss function:
To train the model, we use the cross-entropy loss, which measures the difference between the predicted probability distribution and the true label:\[\mathcal{L} = -\sum_{c=1}^C I(y_n = c) \log p(y_n = c|\mathbf{x}_n; \boldsymbol{\theta})\]where:
\(I(y_n = c)\) indicates whether the true class for the \(n\)-th example is \(c\)
\(p(y_n = c|\mathbf{x}_n; \boldsymbol{\theta})\) is the predicted probability of class \(c\) for the \(n\)-th example
Cross-entropy effectively captures the “distance” between the predicted distribution and the true one by focusing on the log-probability of the correct class
Micro and Macro Averaging#
PrecisionandRecallare two commonly used metrics to assess the performance of a classification modelThe metrics are fairly intuitive with binary classification
When it comes to multi-class classification these metrics need to be tweaked a bit to measure performance of each class
Precision=\(\frac{TP}{TP + FP}\) and Recall=\(\frac{TP}{TP + FN}\)
from sklearn.datasets import load_iris as load_data, make_classification
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import train_test_split
import matplotlib.pyplot as plt
from sklearn.metrics import ConfusionMatrixDisplay
X, y = load_data(return_X_y=True)
# split data into train and test
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=0)
y_pred = LogisticRegression(random_state=0).fit(X_train, y_train).predict(X_test)
target_names = ["A", "B", "C"]
plt.rcParams.update({'font.size': 12, "figure.figsize": (5,3)})
ConfusionMatrixDisplay.from_predictions(y_test, y_pred,
display_labels=target_names)
plt.show()
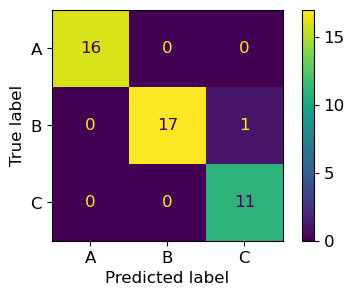
True Positives (TPs) are the metrics on the main diagonal
False Positives (FPs) are the metrics on the columns excluding the ones in the main diagonal, e.g., FPs for class A are cells (2,1) and (3,1)
False Negatives (FNs) are the metrics on the rows excluding the ones in the main diagonal, e.g., FNs for class A are cells (1,2) and (1,3)
Thus, we have
\(TP_A=16, FP_A=0\)
\(TP_B=17, FP_B=0\)
\(TP_C=11, FP_C=1\)
Micro-Averaging: Computes the global (total) counts of true positives (TP), false positives (FP), and false negatives (FN) across all classes
\(\frac{TP_A + TP_B + TP_C} {TP_A + TP_B + TP_C + FP_A + FP_B + FP_C}=\frac{16+17+11}{16+17+11+0+0+1}=\frac{44}{45}=0.977\)
Macro-Averaging: Computes the metric independently for each class and then takes the average\(\text{Precision_A}=\frac{TP_A}{TP_A+FP_A}=\frac{16}{16}=1\)
\(\text{Precision_B}=\frac{TP_B}{TP_B+FP_B}=\frac{17}{17}=1\)
\(\text{Precision_C}=\frac{TP_C}{TP_C+FP_C}=\frac{11}{11+1}=0.92\)
Hence, we have \(\frac{\text{Precision_A}+\text{Precision_B}+\text{Precision_C}}{3}=\frac{2.92}{3}=0.973\)
If the data set is balanced, both micro-average and macro-average will result in similar scores
from sklearn.metrics import classification_report
print(classification_report(y_test, y_pred))
precision recall f1-score support
0 1.00 1.00 1.00 16
1 1.00 0.94 0.97 18
2 0.92 1.00 0.96 11
accuracy 0.98 45
macro avg 0.97 0.98 0.98 45
weighted avg 0.98 0.98 0.98 45
Now let’s see how the micro and macro average scores vary when the dataset is an imbalanced one.
https://scikit-learn.org/stable/modules/generated/sklearn.datasets.make_classification.html
X, y = make_classification(
200,
5,
n_informative=3,
n_classes=3,
class_sep=0.8,
weights=[0.75, 0.1, 0.15],
random_state=0,
)
# split data into train and test
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=14)
y_pred = LogisticRegression(random_state=0).fit(X_train, y_train).predict(X_test)
target_names = ["A", "B", "C"]
import numpy as np
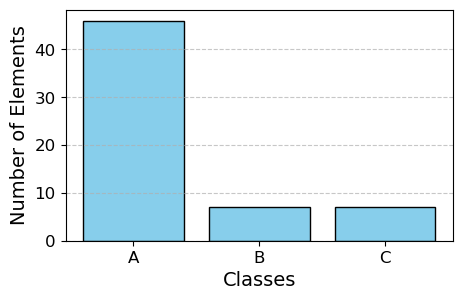
# Count the number of elements in each class
class_counts = np.bincount(y_test)
# Plotting the bar plot
plt.bar(target_names, class_counts, color='skyblue', edgecolor='black')
plt.xlabel('Classes', fontsize=14)
plt.ylabel('Number of Elements', fontsize=14)
plt.xticks(fontsize=12)
plt.yticks(fontsize=12)
plt.grid(axis='y', linestyle='--', alpha=0.7)
plt.show()
plt.rcParams.update({'font.size': 12, "figure.figsize": (5,3)})
ConfusionMatrixDisplay.from_predictions(y_test, y_pred,
display_labels=target_names)
plt.show()
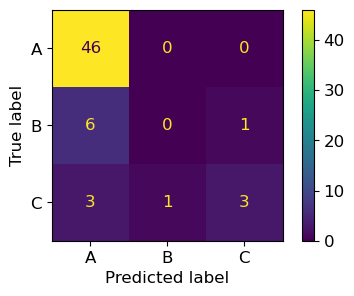
print(classification_report(y_test, y_pred))
precision recall f1-score support
0 0.84 1.00 0.91 46
1 0.00 0.00 0.00 7
2 0.75 0.43 0.55 7
accuracy 0.82 60
macro avg 0.53 0.48 0.49 60
weighted avg 0.73 0.82 0.76 60
46/(46+9)
0.8363636363636363
#macro
(0.84+0+0.75)/3
